Rare Genodermatoses and Use of Next-Generation Sequencing for Evaluation and Diagnosis
July 2025
Dr. Donald Glass presented information about rare genodermatoses and the sequencing techniques used to diagnose them.
First, Glass discussed the strengths and limitations of genetic sequencing techniques, including polymerase chain reaction (PCR) and next-generation sequencing (NGS). PCR requires less genetic material to analyze but can only be used for a limited number of target sequences. It is indicated when testing for v-raf murine sarcoma viral oncogene homolog B1 mutations and T-cell receptor gene rearrangements and to confirm genodermatoses with a clear diagnosis.
NGS analyzes more targets with higher sensitivity than PCR but requires more, higher-quality genetic material. Blood samples are typically used to assess germline involvement and as a control to assess somatic involvement. NGS is appropriate for cancer susceptibility panels, primary immunodeficiency panels, and whole exome sequencing (WES). It is less reliable for detecting deletions, insertions, and splice mutations.
Glass shared information about how dermatologists can order specialized genetic testing. He emphasized that patients need to be informed about the risks and benefits of genetic sequencing before pursuing testing.
Second, Glass described the clinical presentation and diagnostic techniques associated with genodermatoses, including Maffucci syndrome, CDAGS (Craniosynostosis, Delayed closure of the fontanelles, Anal and Genitourinary malformations, and Skin manifestations) syndrome, Von Hippel-Lindau (VHL) syndrome, mosaic overgrowth syndromes, and VEXAS (Vacuoles, E1 enzyme, X-linked, Autoinflammatory, Somatic) syndrome. Maffucci syndrome is caused by postzygotic somatic mutations in the IDH1 or IDH2 genes. It is characterized by vascular malformations (e.g., spindle cell hemangiomas) and enchondromas. Enchondromas become cancerous chondrosarcomas in <10% of cases. Maffucci syndrome can be diagnosed with targeted PCR sequencing of involved skin tissue.
CDAGS syndrome is characterized by craniosynostosis, large open fontanelles, hearing loss, anal anomalies, genitourinary malformations and porokeratosis. Glass described the identification of a causative point mutation on the RNU12 gene using WES of multiple patients and their unaffected parents.
VHL syndrome is characterized by multiple benign and malignant tumors caused by pathogenic variants of the VHL gene. Patients with a history of multiple cancers should be referred to a cancer geneticist, who may ask patients about their cancer history, including breast, ovarian, pancreatic, and renal cancer and assess for appropriate testing (e.g., targeted exome sequencing using multicancer panels). Dermatologists may be asked by cancer geneticists, hematologists, or oncologists to perform punch biopsies when skin tissue is needed as a control sample for NGS.
Mosaic overgrowth syndromes represent a group of disorders characterized by excessive growth in different areas of the body, as the associated genetic changes occur in some cells but not others. They are diagnosed using targeted exome sequencing of normal and involved skin tissue.
VEXAS syndrome is a treatment-refractory inflammatory syndrome that develops in late adulthood and is often fatal. It is characterized by fevers, cytopenia, characteristic vacuoles in myeloid and erythroid precursor cells, dysplastic bone marrow, neutrophilic cutaneous and pulmonary inflammation, chondritis, and vasculitis. VEXAS syndrome is caused by somatic mutations affecting the X-linked UBA1 gene.
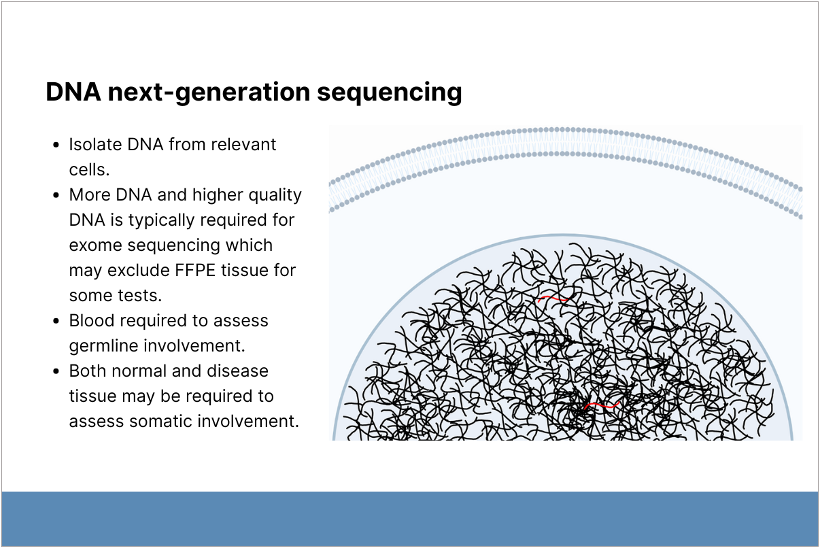
NGS analyzes more targets with higher sensitivity than PCR but requires more, higher-quality genetic material.
Mark January 28-31, 2026 on your calendar for the 2026 Annual DF Clinical Symposium.

